Single Particle Image Processing
We also investigate our samples of interest using elecron microscopy followed by single particle image processing.
Since we are working with single myosin molecules of ~150kDa, our main approach is negative staining. With this method, molecules of such small sizes (and smalle) can be resolved, which was for a long time not possible with cryo EM. Recent research and improvements make it possible to resolve single molecules of sizes far below 200kDa with cryo EM (mainly due to the use of a phase plate). However, the negative stain approach is still a very powerful method, especially when no information about the structure of the molecule is known.
Due to our collaborations we have access to a Philips CM100 and a FEI Tecnai electron microscope at the Dietz Lab (TUM) as well as a JEOL JEM-1011 at the chair of Joachim Rädler (LMU).
For the image processing (e.g. particle picking, alignment and classification) a large number of different softwares is available. Here we mainly use SPIDER (System
number of different softwares is available. Here we mainly use SPIDER (System
for Processing Image Data from Electron microscopy and Related Fields) & WEB. Other software packages such as Scipion, XMIPP or EMAN2 are also applied.
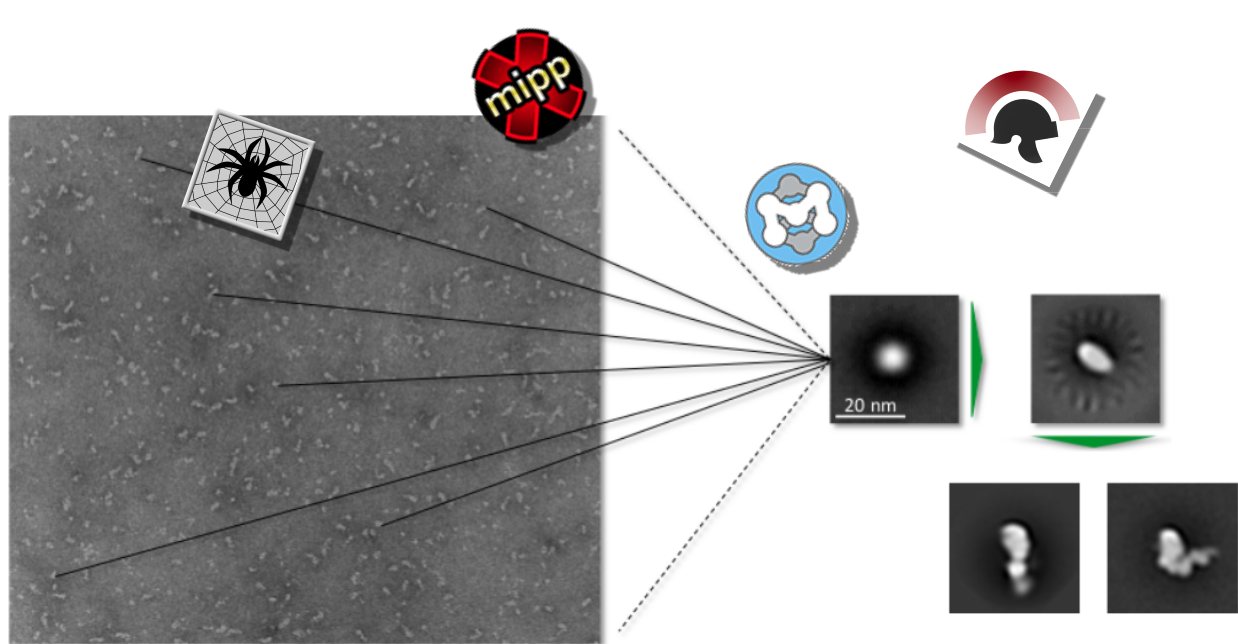
Publications containing single particle image processing data:
- Self-organization of actin networks by a monomeric myosin. Saczko-Brack D, Warchol E, Rogez B, Kröss M, Heissler S M, Sellers J R, Batters C, Veigel C: Proc Natl Acad Sci USA. (2016) Nov 7
- Calcium gets myosin VI ready for word. Masters TA, Kendrick-Jones J, Buss F: Proc Natl Acad Sci USA. Vol.113 (9) (2016) Mar 1 Commentary in PNAS on our PNAS-Paper (2016) Jan 25
- Calmodulin regulates dimerisation, motility and lipid binding of Leishmania myosin XXI. Batters C, Ellrich H, Helbig C, Woodall KA, Hundschell C, Brack D, Veigel C Proc Natl Acad Sci USA Vol. 111 (2): E227-E236 (2014)
- Cloning, expression and characterisation of a novel molecular motor, Leishmania myosin-XXI. Batters C, Woodall KA, Toseland CP, Hundschell C, Veigel C. J Biol Chem 287:27556-66 (2012)
- A monomeric myosin VI with a large working stroke. Lister I, Schmitz S, Walker M, Trinick J, Buss F, Veigel C, Kendrick-Jones J. EMBO J. 23: 1729-38 (2004).
- Myosin VI: cellular functions and motor properties. Roberts R, Lister I, Schmitz S, Walker M, Veigel C, Trinick J, Buss F, Kendrick-Jones J. Philos Trans R Soc Lond B Biol Sci. 359: 1931-44 (2004).

